Personalised Immunotherapy

Our Research: Methods & vaccines 2.0
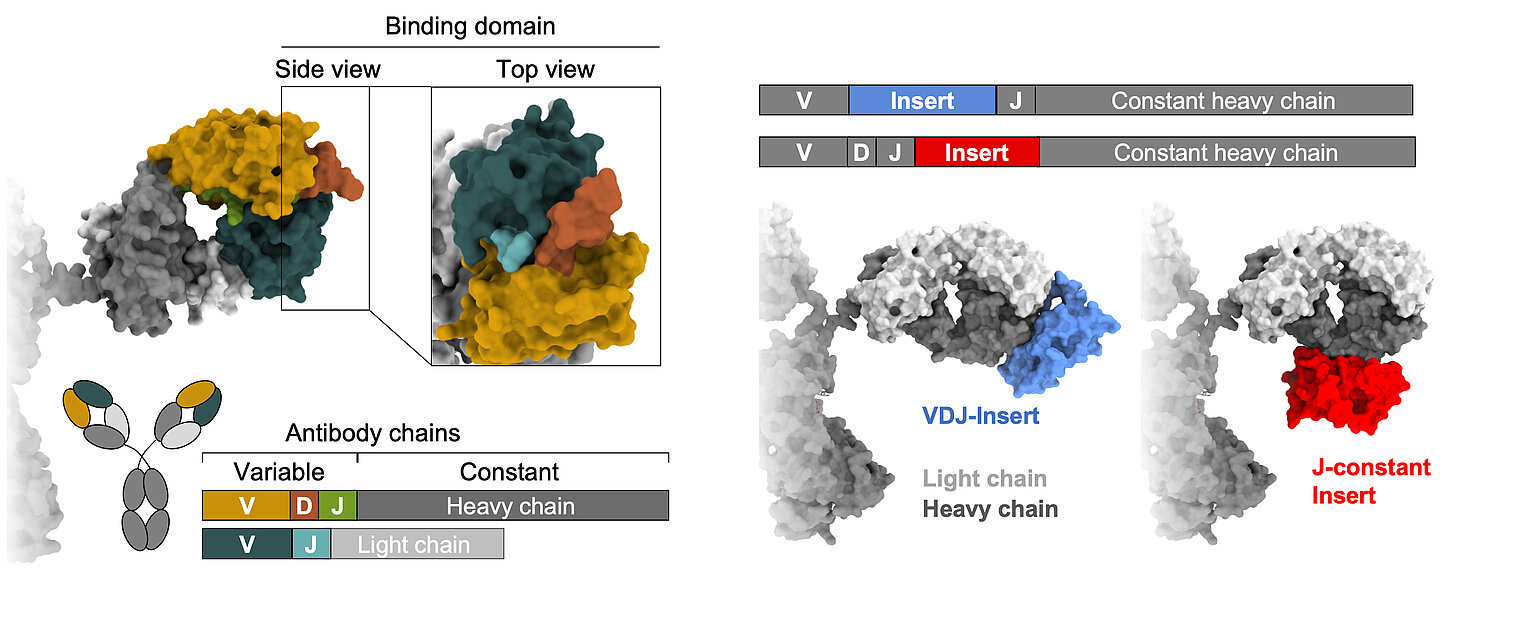
Learning from nature, we strive to gain a comprehensive understanding of B cell diversification and function in humans. In addition to classical recombination events, we study a rare but sometimes particularly useful diversification process: the acquisition of large DNA inserts in the heavy chain genes that equip antibodies with extra domains. Our research is based on three main pillars:
Our first research goal is to develop novel tools to study the repertoire of antibodies and the underlying recombination events. One of our methods holds promise for the personalized assessment of genomic scarring. Prospectively, a biomarker for DNA repair may be used for the prediction and prognosis of diseases related to DNA-repair malfunction, such as defects of the immune system.
Our second goal is to develop personalized cell-based vaccines. After identifying the blind spots in our B cell repertoire, B cells “by design” may extend the natural repertoire through artificial immunity to endow the body with antibodies of superior reactivity.
The third branch of our research arose during the SARS-CoV-2 pandemic. We aim to improve the safety and efficacy of vaccines by preventing their interaction with body-own structures via so-called Body-Inert, but B cell Activating vaccines (BIBAX). In the future, we aim to explore how interactions of vaccines with body receptors hamper immune responses to pathogens.
Our Research: Methods & vaccines 2.0
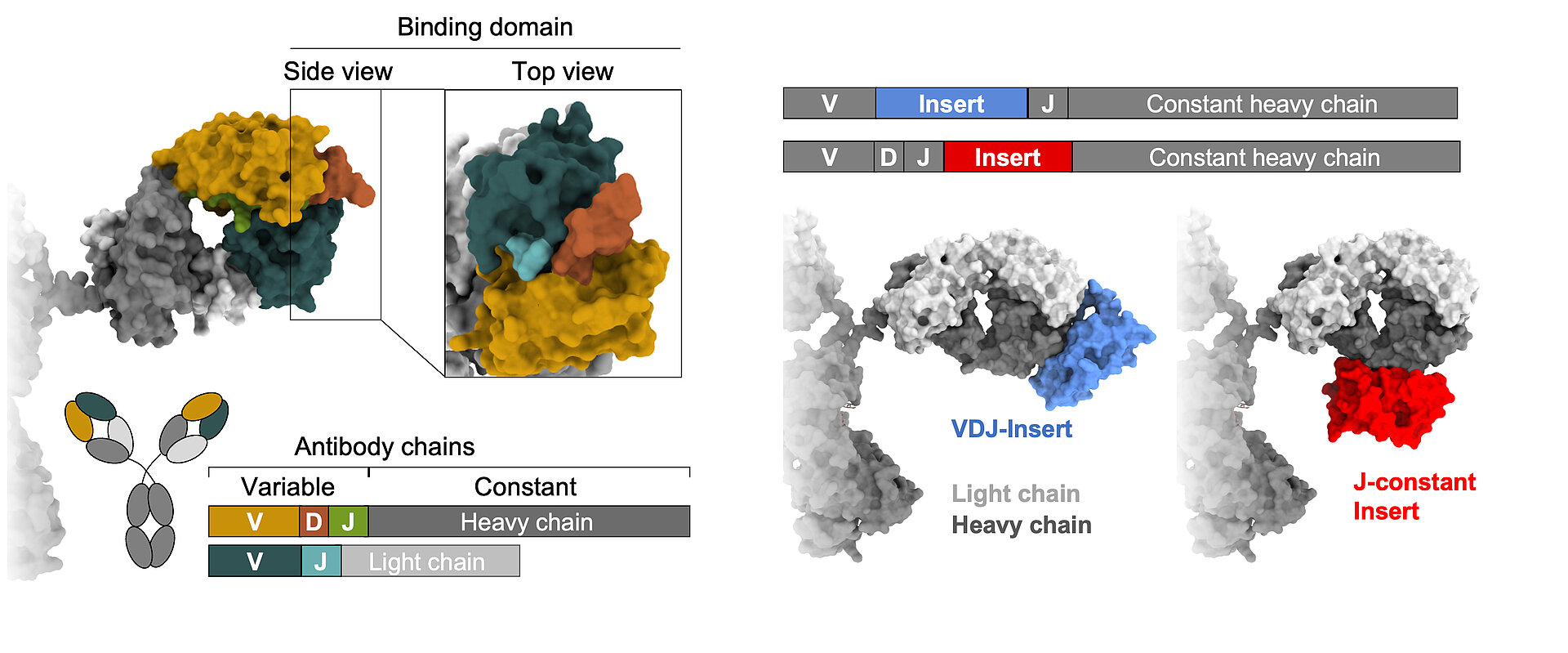
Learning from nature, we strive to gain a comprehensive understanding of B cell diversification and function in humans. In addition to classical recombination events, we study a rare but sometimes particularly useful diversification process: the acquisition of large DNA inserts in the heavy chain genes that equip antibodies with extra domains. Our research is based on three main pillars:
Our first research goal is to develop novel tools to study the repertoire of antibodies and the underlying recombination events. One of our methods holds promise for the personalized assessment of genomic scarring. Prospectively, a biomarker for DNA repair may be used for the prediction and prognosis of diseases related to DNA-repair malfunction, such as defects of the immune system.
Our second goal is to develop personalized cell-based vaccines. After identifying the blind spots in our B cell repertoire, B cells “by design” may extend the natural repertoire through artificial immunity to endow the body with antibodies of superior reactivity.
The third branch of our research arose during the SARS-CoV-2 pandemic. We aim to improve the safety and efficacy of vaccines by preventing their interaction with body-own structures via so-called Body-Inert, but B cell Activating vaccines (BIBAX). In the future, we aim to explore how interactions of vaccines with body receptors hamper immune responses to pathogens.
Prof Dr Kathrin de la Rosa
Our mission is to understand the universe of the antibody repertoire. Our vision is to transform this knowledge into personalized solutions for infectious diseases.

Kathrin de la Rosa (born Kathrin Pieper in 1984) became an immunologist after completing her doctoral studies in 2013 at the University Medical Center Freiburg on B cell disorders in immunodeficient patients. As a postdoctoral fellow, she decided to study monoclonal antibodies in infectious diseases. To this end, she joined the research group of Antonio Lanzavecchia at the Institute for Research in Biomedicine in Switzerland. In 2018, she started her own research group at the Max Delbrück Center in the Helmholtz Association (MDC) in Berlin after being awarding the Emmy Noether Fellowship of the German Research Foundation (Deutsche Forschungsgemeinschaft, DFG). In 2020, she was awarded the European Research Council (ERC) starting grant and was selected to obtain the Johanna Quandt Fellowship of the Stiftung Charité. She became a W2 Professor of “Immune mechanisms in translation” at the Berlin Institute of Health@Charité in 2021 and, since 2024, is a W3 Professor of Personalized Immunotherapy (W3) at the Hannover Medical School (MHH) in a joint appointment with the Helmholtz Centre for Infection Research (HZI). She is running her research group at the Centre for Individualised Infection Medicine (CiiM).









Selected Publications
Lebedin, M., C. Ratswohl, A. Garg, M. Schips, C. V. García, L. Spatt, C. Thibeault, B. Obermayer, J. Weiner, I. M. Velásquez, C. Gerhard, P. Stubbemann, L.-G. Hanitsch, T. Pischon, M. Witzenrath, L. E. Sander, F. Kurth, M. Meyer-Hermann*, and K. de la Rosa*. 2024. Soluble ACE2 correlates with severe COVID-19 and can impair antibody responses. iScience 27: 109330. DOI: 10.1016/j.isci.2024.109330. PMID: 38496296. Open Access
Lebedin, M., C. V. García, L. Spatt, C. Ratswohl, C. Thibeault, L. Ostendorf, T. Alexander, F. Paul, L. E. Sander, F. Kurth, and K. de la Rosa. 2023. Discriminating promiscuous from target‐specific autoantibodies in COVID‐19. Eur. J. Immunol. 53: e2250210. DOI: 10.1002/eji.202250210. PMID: 36856018. Open Access
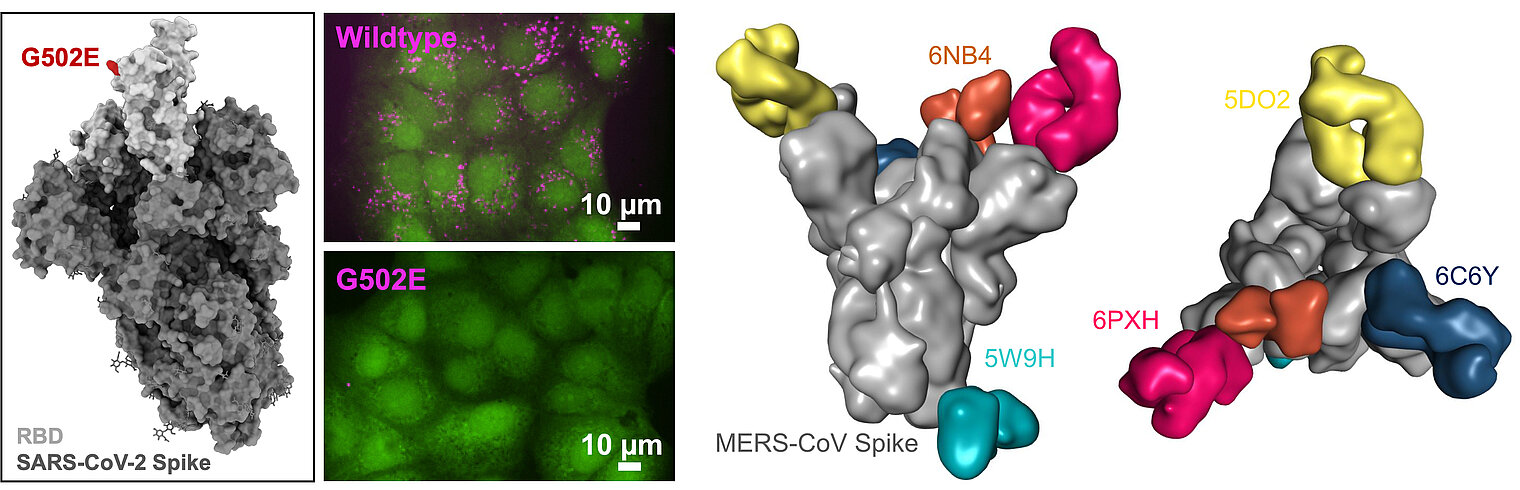
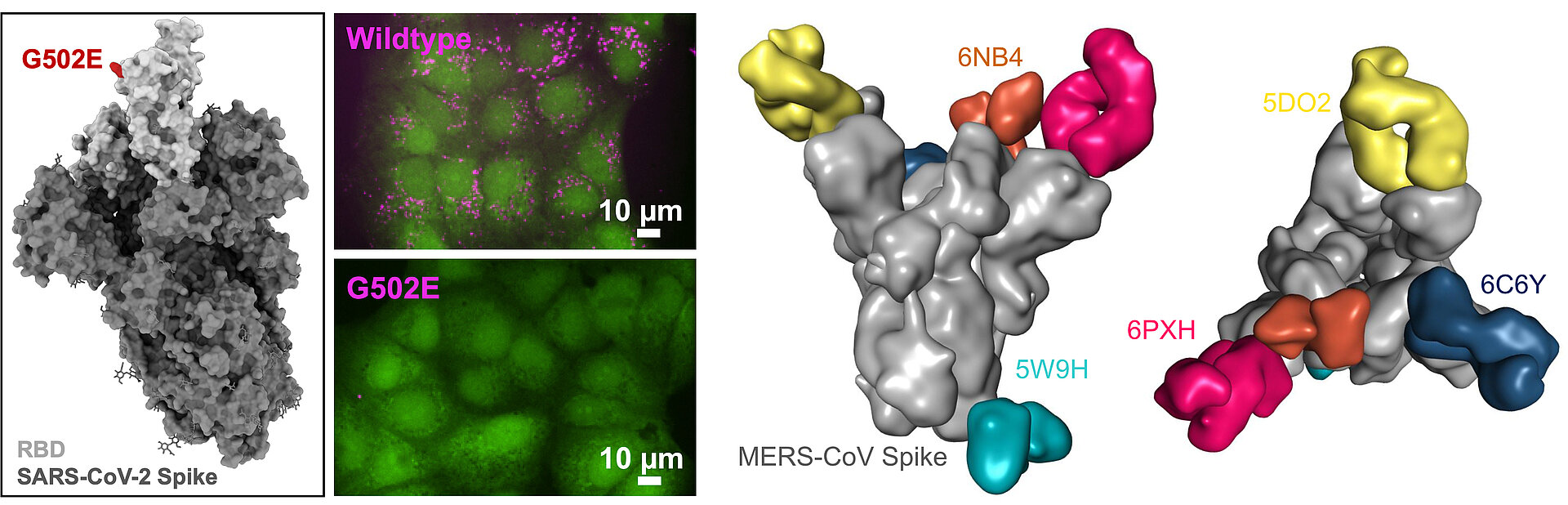
Ratswohl, C., C. V. García, A. ul W. Ahmad, H. Gonschior, M. Lebedin, C. E. Silvis, L. Spatt, C. Gerhard, M. Lehmann, L. E. Sander, F. Kurth, S. Olsson, and K. de la Rosa. 2023. A design strategy to generate a SARS‐CoV‐2 RBD vaccine that abrogates ACE2 binding and improves neutralizing antibody responses. Eur. J. Immunol. 53: e2350408. DOI: 10.1002/eji.202350408. PMID: 37435628. Open Access
Lebedin, M., M. Foglierini, S. Khorkova, C. V. García, C. Ratswohl, A. N. Davydov, M. A. Turchaninova, C. Daubenberger, D. M. Chudakov, A. Lanzavecchia, and K. de la Rosa. 2022. Different classes of genomic inserts contribute to human antibody diversity. Proc National Acad Sci 119: e2205470119. DOI: 10.1073/pnas.2205470119. PMID: 36037353. Open Access
Pieper, K.*, J. Tan*, L. Piccoli*, M. Foglierini, S. Barbieri, Y. Chen, C. Silacci-Fregni, T. Wolf, D. Jarrossay, M. Anderle, A. Abdi, F. M. Ndungu, O. K. Doumbo, B. Traore, T. M. Tran, S. Jongo, I. Zenklusen, P. D. Crompton, C. Daubenberger, P. C. Bull, F. Sallusto, and A. Lanzavecchia. 2017. Public antibodies to malaria antigens generated by two LAIR1 insertion modalities. Nature 548: 597–601. DOI: 10.1038/nature23670. PMID: 28847005
Publications
A complete list of publications can be found in the ORCID profile of Kathrin de la Rosa.