Computational Biology for Individualised Medicine

Research focus
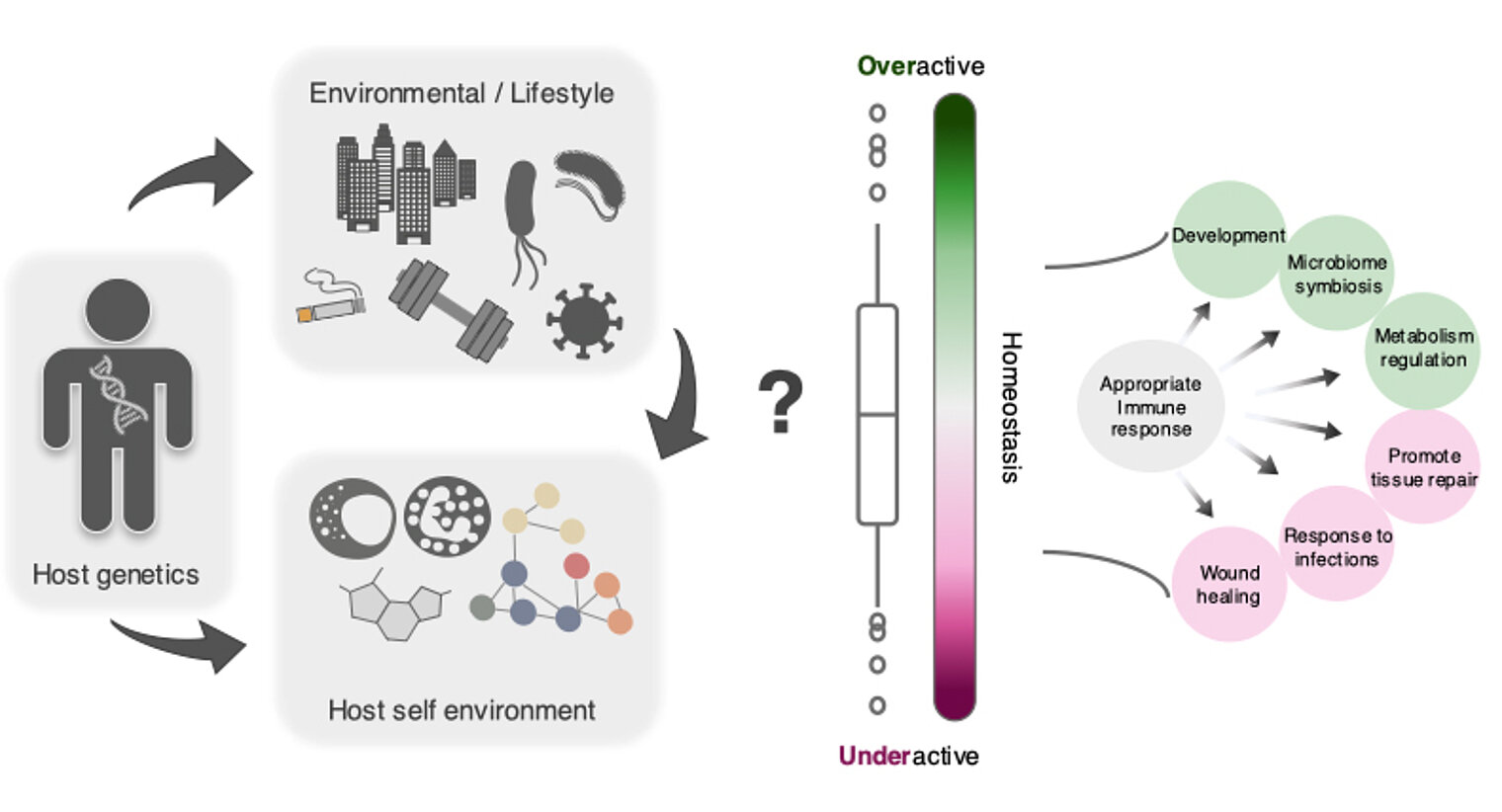
The research group “Computational Biology for Individualized Infection Medicine” studies the interaction of genetic background and environment, and its contribution to infection and immune-related diseases.
We focus on applying and developing the computational and statistical approaches to study the effect of genetic factors on a variety of molecular levels (such as genetics, genomics, metabolomics etc.), immunological parameters and functions, and complex diseases.
Our aim is to reveal the host genetic risk factors and their downstream molecular pathways as well as to improve the identification of at-risk patients, which are crucial to make progress in understanding and treating infectious diseases in an individualized manner.

Research focus
The research group “Computational Biology for Individualized Infection Medicine” studies the interaction of genetic background and environment, and its contribution to infection and immune-related diseases.
We focus on applying and developing the computational and statistical approaches to study the effect of genetic factors on a variety of molecular levels (such as genetics, genomics, metabolomics etc.), immunological parameters and functions, and complex diseases.
Our aim is to reveal the host genetic risk factors and their downstream molecular pathways as well as to improve the identification of at-risk patients, which are crucial to make progress in understanding and treating infectious diseases in an individualized manner.
Prof. Dr. Yang Li
People are different in genetic background. We study this inter-individual variation and want to understand susceptibility to infections and predict disease risk

Yang Li is a computational biologist focusing on integration of multi-omics and single cell omics data to study the interaction between genetic background and the environment, and its contribution to infection and immune-related diseases. Y. Li has demonstrated how genetic risk factors of the host and their downstream molecular pathways determine and predict the human immune functions, which are crucial for treating infectious and allergic diseases in an individualized manner (Cell 2016, Nat Med 2016, Nat Immunol 2018). Recently, her team conducted the first multi-omics study of dysfunctional immune system in mild and severe COVID19 patients (Cell 2020) and revealed the epigenetic and genetic regulators of innate immunity in COVID-19 (Cell Genomics 2023). For her research, she received the prestigious ERC Starting Grant “ModVaccine” on improving vaccine efficiency (2020).
Prof. Dr. Li is an Editorial Board Member of several prestigious scientific magazines such as Frontiers in Immunology and Frontiers in Genetics. She is also involved into National and International Reviewer activities by the German Society of Research (DFG), German Center for Infection Research (DZIF), Israel Science Foundation, Swiss National Science Foundation, Medical Research Council (UK) and French Foundation for Medical Research. Her other professional memberships include German Society of Immunology, American Human Genetics Society member and European Human Genetics Society. Prof. Dr. Li is also an Advisory Board Member and Member of Pool of Experts at the Milieu Intérieur consortium, Pasteur Institute, France.
Research projects
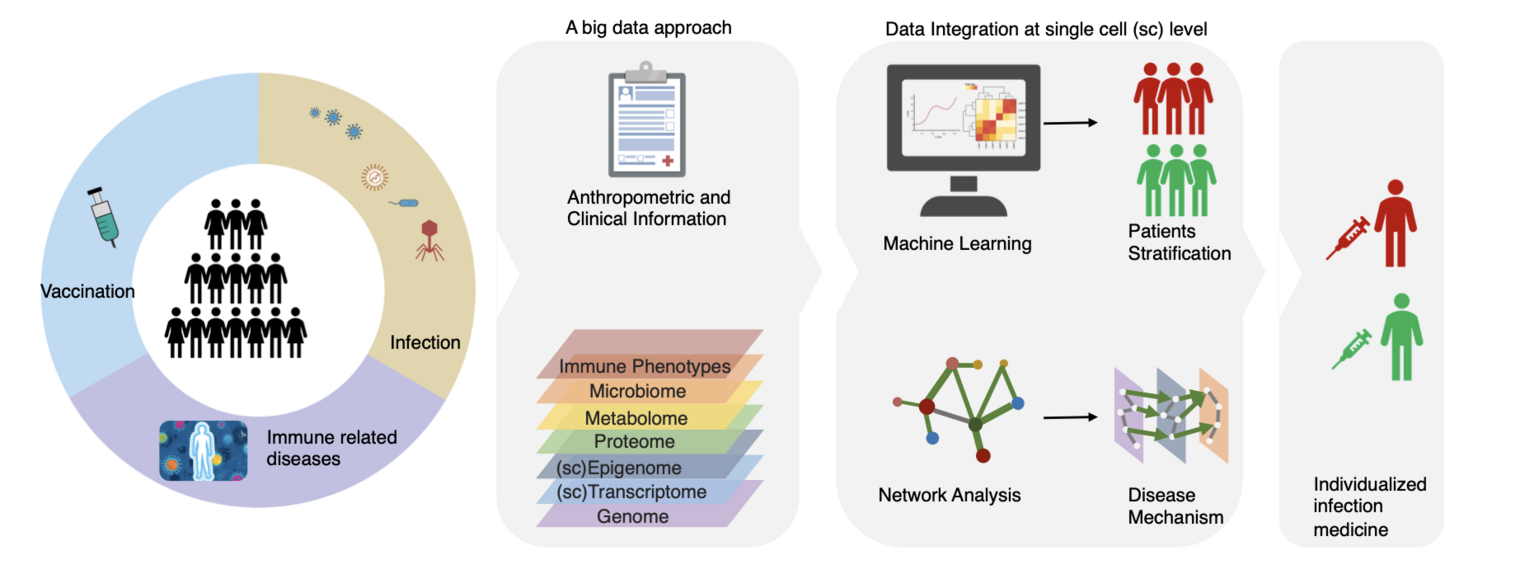
We employ a big data approach to study immune functions in vaccination and infection. Specifically, we focus on the following directions:
Integration of multi-omics data
Our approach involves integrating and analysing multi-omics datasets and immune profiling of patient/control cohorts to develop computational models for detecting disease/trait-associated markers, key biological pathways, and molecular interactions. By doing so, we aim to understand the genetic basis of complex traits and develop individualized medicine.
Single-cell genomics
We also develop and apply cutting-edge techniques to investigate the heterogeneity of cell populations, identify rare cell types, and understand cellular dynamics in various diseases at a single-cellresolution. These techniques can also be applied to study the genetic and epigenetic regulation of gene expression, as well as the interaction between cells and their environment.
Genetic regulation of molecular and immune phenotype
Our research focuses on understanding how genetic variation affects molecular and immune phenotypes, such as gene expression, metabolites, and cytokine responses to stimulations. To achieve this, we develop computational methods and algorithms to fully exploit high-throughput datasets from the most recent profiling technologies, such as causal inference and deconvolution1.
Overall, our big data approach enables us to gain a comprehensive understanding of immune functions in vaccination and infection. By studying the genetic basis and molecular interactions underlying complex traits and diseases, we hope to develop personalized infection medicine that accounts for individual differences in genetic variation and immune responses.
During the last years, our research has been contributing to the following four major domains:
Infectious diseases
Utilizing transcriptome, genomics and epigenome data from the hospitalized and convalescent COVID-19 patients, we demonstrated that an altered chromatin accessibility and allele-specific open chromatin are associated with impaired epigenetic regulation that contributes to COVID-19 pathogenesis (Cell Genomics 2023). By employing single cell multi-omics approaches, we showed minor differences in epigenetic and transcriptional programs of the immune system of post COVID-19 patients, and these changes largely recover to the normal level (Frontiers in Immunol. 2022). However, the circulating proteome is affected in post-COVID-19 patients even several weeks after infection (Frontiers in Immunol. 2022).
Immune-related diseases
By studying dynamic transcriptional changes in single cells of pediatric individuals, we were able to elucidate celiac disease-associated differentially expressed genes and genetic risk loci (Frontiers in Immunol. 2022). Furthermore, we performed a functional genomics study in T1D and revealed novel insights into the genetic factors that affect immunological responses in T1D (eLife 2022).
Together with clinicians at the MHH, we performed a single cell multi-omics study of atopic dermatitis (AD) and psoriasis (PS) and identified that the disease-specific T cell clusters were mostly of a Th2/Th22 sub-population in AD and Th17/Tc17 in PS and their numbers were associated with severity scores in both diseases (Allergy 2022).
Vaccine response
Using an integrative genomics approach in two independent population-based BCG vaccination cohorts of healthy individuals, we detected genetic loci that influence cytokine responses of trained immunity and the H3K9 histone demethylases of the KDM4 family as major regulators of trained immunity responses (Eur. J. Immunol. 2022). Next, we presented a comprehensive view of cellular transcriptional programs of trained immunity at a single-cell resolution, which outlines the heterogeneity in gene expression in trained cells (J. Clin. Invest 2022). We also observed that circulating metabolites are important factors influencing BCG-induced trained immunity in humans and modulating metabolic pathways may be a novel strategy to improve vaccine and trained immunity responses (PLOS Biol 2022).
Development of novel computational methods
We demonstrated that eQTL deconvolution analyses can be a powerful tool to understand cell-type-specific molecular mechanisms of asthma development (Allergy 2022).












Selected publications
1. Chu, X., Janssen, A.W.M., Koenen, H., Chang, L., He, X., Joosten, I., Stienstra, R., Kuijpers, Y., Wijmenga, C., Xu, C.- J., Netea, M.G, Tack, C.J., Li, Y.# A genome-wide functional genomics approach uncovers genetic determinants of immune phenotypes in type 1 diabetes. eLife, 11, e73709 (2022). https://doi.org/10.7554/eLife.73709
2. Koeken, V.A.C.M., Qi, C., Mourits, V.P., de Bree, L.C.J., Moorlag, S.J.C.F.M., Sonawane, V., Heidi Lemmers, Dijkstra, H., Joosten, L.A.B., van Laarhoven, A., Xu, C.-J., van Crevel, R., Netea, M.G., Li, Y.# Plasma metabolome predicts trained immunity responses after antituberculosis BCG vaccination. PLoS Biol20(9): e3001765, (2022). https://doi.org/10.1371/journal.pbio.3001765
3. Qi, C., Berg, M., Chu, X., Van Den Berge, M., Xu, C., Koppelman, G., Nawijn, M., Li, Y. Cell type eQTL deconvolution of bronchial epithelium through integration of single cell and bulk RNA-seq. Allergy 77, 3663-3666 (2022).
https://doi.org/10.1111/all.15410
4. Zhang, B., Moorlag, S., Domínguez-Andrés, J., Bulut, Ö., Kilic, G., Liu, Z., van Crevel, R., Xu,C.-J., Joosten, L.A.B., Netea, M.G.#, Li, Y.# Single-cell RNA sequencing reveals induction of distinct trained immunity programs in human monocytes. J Clin Invest, 132(7), e147719 (2022) https://doi:10.1172/JCI147719
5. Zhang, B, Roesner, L.M., Traidl, S., Koeken, V.A.C.M., Xu, C.-J., Werfel, T., Li, Y. Single-cell profiles reveal distinctive immune response in atopic dermatitis in contrast to psoriasis. Allergy, 00: 1- 15 (2022). https://doi: 10.1111/all.15486
A complete list of Yang Li's publications can be found on GoogleScholar.
Jobs
The Research Department “Computational Biology for Individualised Medicine” welcomes applications from candidates qualified for either a PhD or Postdoc position.
You can find all open job advertisements on the HZI's job page.
Unsolicited applications are always welcome. We are looking for motivated applicants with a strong background in Bioinformatics / Computational Biology, Computer Science, Statistics, Biology or Physics, good programing skills and interest in interdisciplinary research in biology and infection research.
We continuously offer Masters/Bachelor research projects. Please contact us for details by email.
Our research focuses on understanding inter-individual variation in susceptibility to immune related diseases and predicting immune functions through integration of large multi-omics and in-depth immune phenotypes datasets from a variety of existing and planned patient cohorts. Methods that we employ or develop in our research are related to the fields of Bioinformatics, Systems Genetics, Machine learning & deep learning and Systems immunology.
Unless otherwise stated in a specific job advertisement, please send your applications to Yang Li. Please note our data protection statement.
Further information
Development of novel computational methods
Since studies based on bulk RNA-seq data are constrained by the lack of cell-type resolution, we developed an eQTL deconvolution approach to identify cell-type specific genetic effect of gene expression of disease-associated variants (BMC Bioinfo. 2020). Our results were replicated by single-cell eQTL method, thereby suggesting eQTL deconvolution analyses can be a powerful tool to understand cell-type-specific molecular mechanisms of asthma development (Allergy 2022). A demultiplexing analysis framework has been developed in my lab, which will streamline the future analysis of single-cell omics datasets (Zoodsma et al., in preparation).
Further details and models can be found here.